SciPy v. Octave - Round one
June 17, 2012 at 10:55 PM by Dr. Drang
On Friday, while working out a sampling plan for a client, I needed to calculate the 95% one-sided lower confidence limits for binomial sampling with a small number of samples.1 I figured it’d be a good little project for comparing Octave, which I’ve been using for years, and SciPy, which I’ve been promising myself I’d learn.
The idea behind binomial sampling is simple. You run a set of pass/fail trials on samples taken from the full population and count the number of passes and fails. With as the number of trials and as the number of failures, your estimate of the probability of failure within the entire population, , is
To figure out how good this estimate is, you calculate confidence limits. In the situation I was dealing with on Friday, I cared only about the lower confidence limit. After the trials were done, I wanted to be 95% confident that was greater than some value.
If you have lots of trials, the confidence limit calculation is pretty easy because you can take advantage of the central limit theorem and assume your estimate of follows a normal distribution. Unfortunately, I wasn’t in that situation—the client wanted to know what could be learned about the probability of failure from a small number of samples.
Here’s how it’s calculated. Solve the nonlinear equation,
for . That will be the 95% one-sided lower confidence limit for .2
Because I was designing a sampling plan, I needed to calculate the lower confidence limits for several combinations of and . I started out with my old friend, Octave:
1: for n = 3:9
2: for y = 0:(n-1)
3: p = fsolve(@(q) binocdf(y, n, q) - .95, (y + .1)/n);
4: printf("%6.4f ", p)
5: endfor
6: printf("\n")
7: endfor
This gave me a nice table of results:
0.0170 0.1354 0.3684
0.0127 0.0976 0.2486 0.4729
0.0102 0.0764 0.1893 0.3426 0.5493
0.0085 0.0628 0.1532 0.2713 0.4182 0.6070
0.0073 0.0534 0.1288 0.2253 0.3413 0.4793 0.6518
0.0064 0.0464 0.1111 0.1929 0.2892 0.4003 0.5293 0.6877
0.0057 0.0410 0.0977 0.1688 0.2514 0.3449 0.4504 0.5709 0.7169
The rows of the table correspond to values of 3 through 9, and the columns correspond to values of 1 through 9. I didn’t bother calculating the values for because they’d all be zero. The upper right triangle of the table is blank because can’t be greater than (we can’t have more failures than trials).
The binocdf function does the work of the summation in the formula above. Because I have the variable y looping from 0 to n-1, it’s acting as in the formula.
The fsolve function is Octave’s nonlinear equation-solving routine. Its two arguments are the function to find the zeros of and the starting guess. The function is defined inline using Octave’s anonymous function syntax, which starts with @. Each time through the loops on n and y, fsolve finds the value of q for which
binocdf(y, n, q) - .95
is zero.
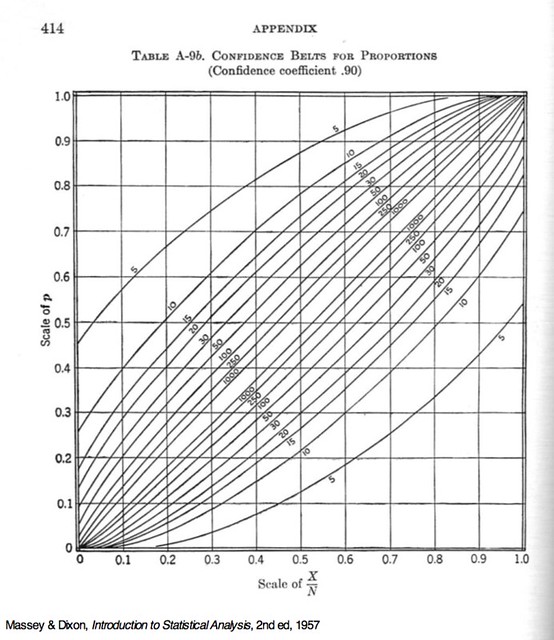
The initial value, (y + .1)/n, was chosen by looking at this chart, from Massey and Dixon’s Introduction to Statistical Analysis.
The lower confidence limits I want for can be read off the lower curve at the (our ) values of 0.2, 0.4, 0.6, 0.8, and 1.0, so I had a decent idea of where to start for set of solutions. The formula I chose didn’t give the most accurate initial guesses in the world,
| x | y | Initial guess | Final value |
|---|---|---|---|
| 1 | 0 | 0.02 | 0.0102 |
| 2 | 1 | 0.22 | 0.0764 |
| 3 | 2 | 0.42 | 0.1893 |
| 4 | 3 | 0.62 | 0.3426 |
| 5 | 4 | 0.82 | 0.5493 |
but it was simple and provided starting values close enough for convergence, which is all that matters.
I used Octave first because I needed to be confident3 that I would get the answers. I knew pretty much how I was going to proceed before starting; the only things I had to look up in the documentation were the syntax for anonymous functions, which I’d never used before, and the printf function, which I just just needed to confirm the existence of—it works like every other printf I’ve seen.
With the real work done, I could relax and play around with SciPy. I Googled “scipy binomial cdf” and came up with this page in the docs. Then I searched for “scipy nonlinear solver” and came up with two likely answers: the fsolve solver, which I believe works the same way the Octave solver of the same name works; and the newton solver, which, despite its name, uses the secant method if a derivative function isn’t supplied. Because the newton solver was designed for scalar functions and fsolve was designed for vectors, I figured newton would be the more appropriate choice. Given how little computation was necessary, I doubt my choice made much difference.
The Python script looked like this:
python:
1: from scipy.stats import binom
2: from scipy.optimize import newton
3:
4: for n in range(3,10):
5: for y in range(n):
6: p = newton(lambda q: binom.cdf(y, n, q) - .95, (y + .1)/n)
7: print "%6.4f " % p,
8: print
It gave the same results as the Octave script.
Apart from the import statements, the form of the script is the same as that of the Octave solution, which is encouraging. Also encouraging was how quickly I was able to find the functions I needed and how easy the documentation was to read.
Not so encouraging were the two warnings Python spit out when I ran the script:
/Library/Python/2.7/site-packages/scipy-0.10.1-py2.7-macosx-10.7-x86_64.egg/
scipy/stats/distributions.py:30: RuntimeWarning: numpy.ndarray size changed,
may indicate binary incompatibility
import vonmises_cython
/Library/Python/2.7/site-packages/scipy-0.10.1-py2.7-macosx-10.7-x86_64.egg/
scipy/stats/distributions.py:30: RuntimeWarning: numpy.ufunc size changed,
may indicate binary incompatibility
import vonmises_cython
The warnings were generated by Line 1.
Since I’m pretty sure NumPy and SciPy were compiled locally when I installed them, I don’t know how or why there’d be a binary incompatibility. I suppose I should Google these warning messages to see if there’s a fix. As I said, I got the right answers with SciPy, but I know that only because I did the problem with Octave first. I’d like to be a little more certain that SciPy is working before I hand all my numerical work over to it.